Jacob Scott Laboratory
-
Jacob Scott Laboratory
- Principal Investigator
- Research
- Our Team
- Publications
- Careers
- Research News

Jacob Scott, MD, DPhil
Staff, Translational Hematology & Oncology Research; Radiation Oncology
Associate Professor, School of Medicine, Case Western Reserve University
Associate Director for Data Science, Case Comprehensive Cancer Center
Adjunct Associate Professor, Department of Physics, Case Western Reserve University
Location: Cleveland Clinic Main Campus
Research
Cancer is a complex disease that is an aberration of our own tissues, but it still obeys fundamental biological rules. Our greatest challenge in the clinic is the emergence of resistance to our therapies, a process which is governed by Darwinian evolution. Using a suite of mathematical and experimental models, my laboratory seeks to deconvolute the complexity of the evolutionary process into fundamental principles. We aim to use this knowledge to then curtail the evolutionary process to increase the efficacy of targeted therapies and radiation. This same knowledge can be further harnessed to understand the differences in disease progression and therapy response on a personalized basis, so that the right treatment can be given to the right patient at the right time.
Biography
I am a veteran of the US Navy submarine force turned academic physician-scientist. Our lab pursues research decomposing the complexity of cancer through mathematical modeling and the biological and clinical validation of these models. My educational background in physics, medicine, mathematics and engineering gives me a unique perspective on cancer and systems biology and I am able to communicate and collaborate with professionals across many disciplines. I have worked extensively on mathematical modeling of cancer evolution and treatment using a variety of models including evolutionary game theory, cellular automata, differential equations and Markov chains. My DPhil thesis focused on the role of heterogeneity, both genetic and microenvironmental, on cancer evolution and radiation response, and my laboratory’s focus is cancer evolution and therapy resistance. Since starting Theory Division, we have begun to diversify, and the lab now has a significant experimental component, conducting experimental evolution in cancer cell lines as well as bacteria. The combination of mathematics, experimental evolution and a clinical focus makes our laboratory stand out as one of the most interdisciplinary in the field of translational cancer evolution and evolutionary medicine. I am eager to use my distinct perspective to help advance this field to help cancer patients.
Education & Professional Highlights
Appointed
2016
Education & Fellowships
Graduate School - St. Anne's College Oxford University
Oxford, OX2, United Kingdom
2018
Residency - H. Lee Moffitt Cancer Center
Radiation Oncology
Tampa, FL USA
2015
Internship - University Hospitals of Cleveland Health System
Internal Medicine
Cleveland, OH USA
2009
Medical Education - Case Western Reserve University
Cleveland, OH USA
2008
Graduate School - Old Dominion University
Engineering Management
Norfolk, VA USA
2003
Undergraduate - United States Naval Academy
Physics
Annapolis, MD USA
1998
Certifications
Radiology - Radiation Oncology
Research
Overview
Theory Division
We now know that cancer proceeds via combined evolutionary and ecological processes. There is irrefutable evidence that populations of cancer cells change as individual cells mutate and selection forces act to shape the population into more fit phenotypes. We now understand that not only do individual cells escape these selection forces by interacting with their environment, including stromal cells like fibroblasts, but also with other tumor cells. However, while basic scientists have established these facts, little, if any, of this knowledge has translated into clinically actionable information; and, more frustratingly, while many scientists have begun to accept these facts, it has not changed the pursuit of ‘silver bullets’: drugs to act on single targets awash in a sea of heterogeneity. In Theory Division, we use a host of tools to approach this problem, with our focus on the evolutionary mechanisms themselves, rather than the specific solutions evolution finds. Below is a snapshot of a few of the things we like to think about... please reach out with questions or ideas - science is a team sport!
Evolution On Adaptive (Fitness) Landscapes
Evolution can be described in many ways. One mathematicall convenient way is consider evolution like a search for peaks (points of highest fitness) on a rugged landscape. If you allow each unique genotype to be a location in space (say the x,y coordinates on a map), then the phenotype is the 'height'. If you now consider each 'map' to be a different drug, you start to reason about how different therapies would affect populations under selection by them. We have modelled bacterial evolution in this way and found that evolution could be effectively 'steered', by a clever ordering of different landscapes, though in reality it is not perfectly controllable because of evolutionary contingencies, but may be predictable. More recently, we have worked to incorporate machinery from statistical mechanics, together with the Hinczewski group in Case Physics to formally derive counter-diabatic driving protocols to allow precise control of the speed and trajectory of populations as they navigate these landscapes.
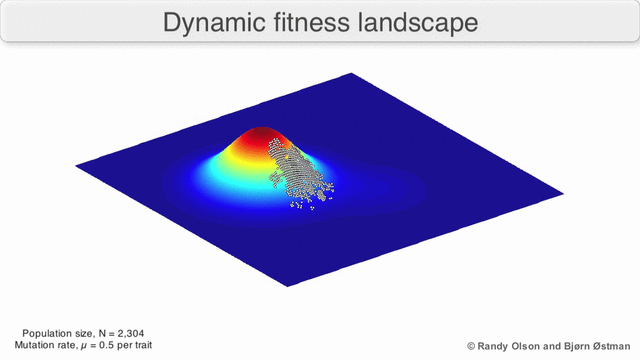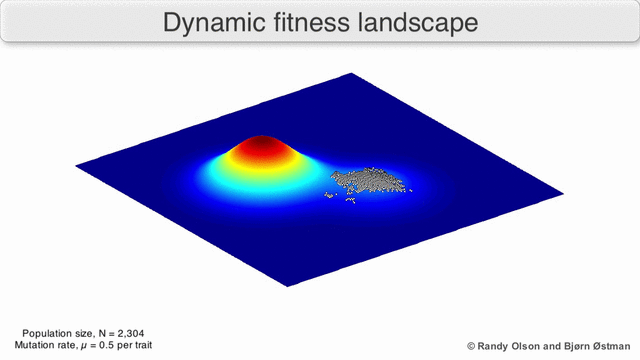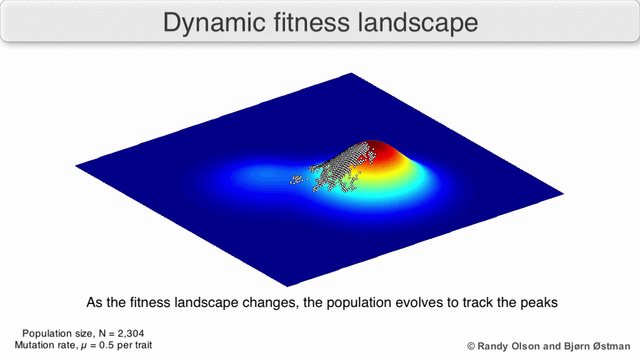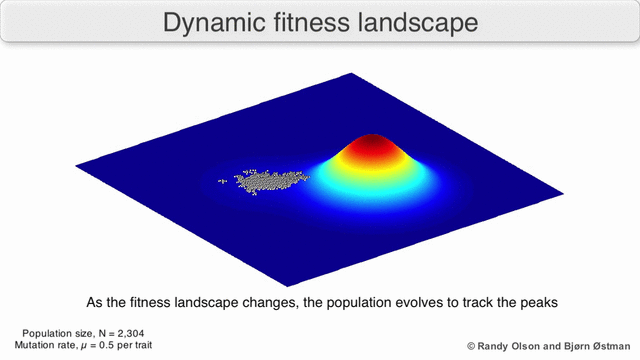
Olson R. and Østman B., "Using fitness landscapes to visualize evolution in action."
Here we show evolution proceeding on three different landscapes in a row. At time 20k and 40k the drugs (landscapes) are switched. In this case, we show an example where the middle drug, cefprozil, is used to steer the evolution of the third drug, ampicillin, to a suboptimal peak. Details can be found here.
Evolutionary Bioreactor
The emergence of drug resistance is the key stumbling block in our fight against cancer. Though our tools can be extremely effective in the short term, it is inevitable that they willfail in the majority of cancers. As an evolutionary phenomenon, we know that the cancer we are treating will “figure out” a way to evade, subvert, and fight back against what we throw at it. We know where the cancer will end up but have relatively little idea how it gets there. We are designing and building a device based on the “morbidostat” framework from the study of antibiotic resistance which we call the EVE system (EVolutionary biorEactor). This will allow us to watch and measure cancer cells on their evolutionary journey towards resistance. It will subject the cells to the same therapies they would experience in patients, but allow us to observe how the cells grow and change much more closely than we ever could in people. The system will be fully robotically automated, providing us a tightly-controlled experimental environment where we can manipulate conditions and see how the cancer cells adjust. Information about the paths that cancers take to resistance can give us new insights about when and how we can intervene while we still can. Our device will allow us to ask and answer questions such as “Can we steer a cancer’s path to slow its progression to resistance?” and “Are there certain times that are better to act than others?”
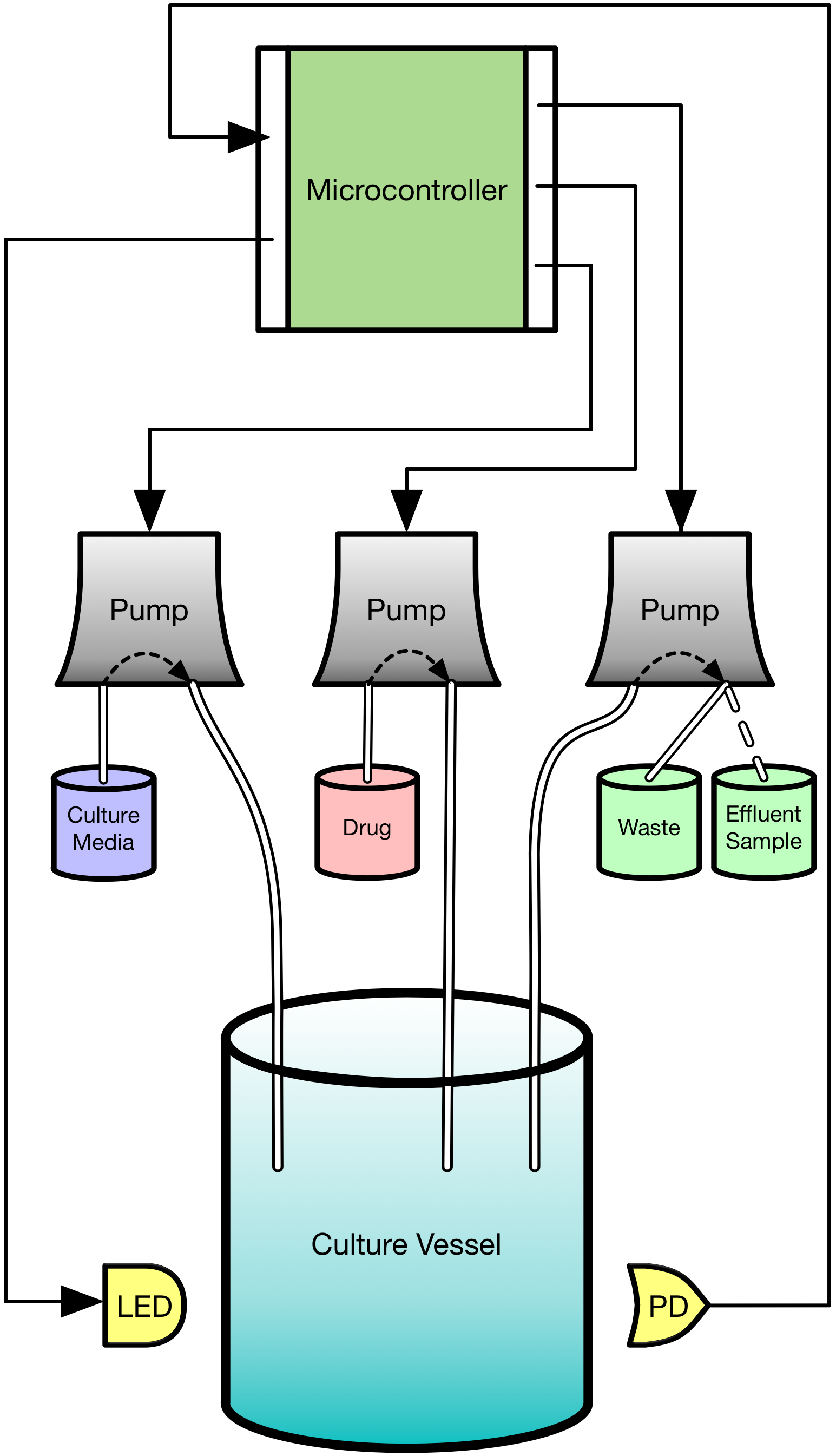
Schematic of evolutionary bioreactor
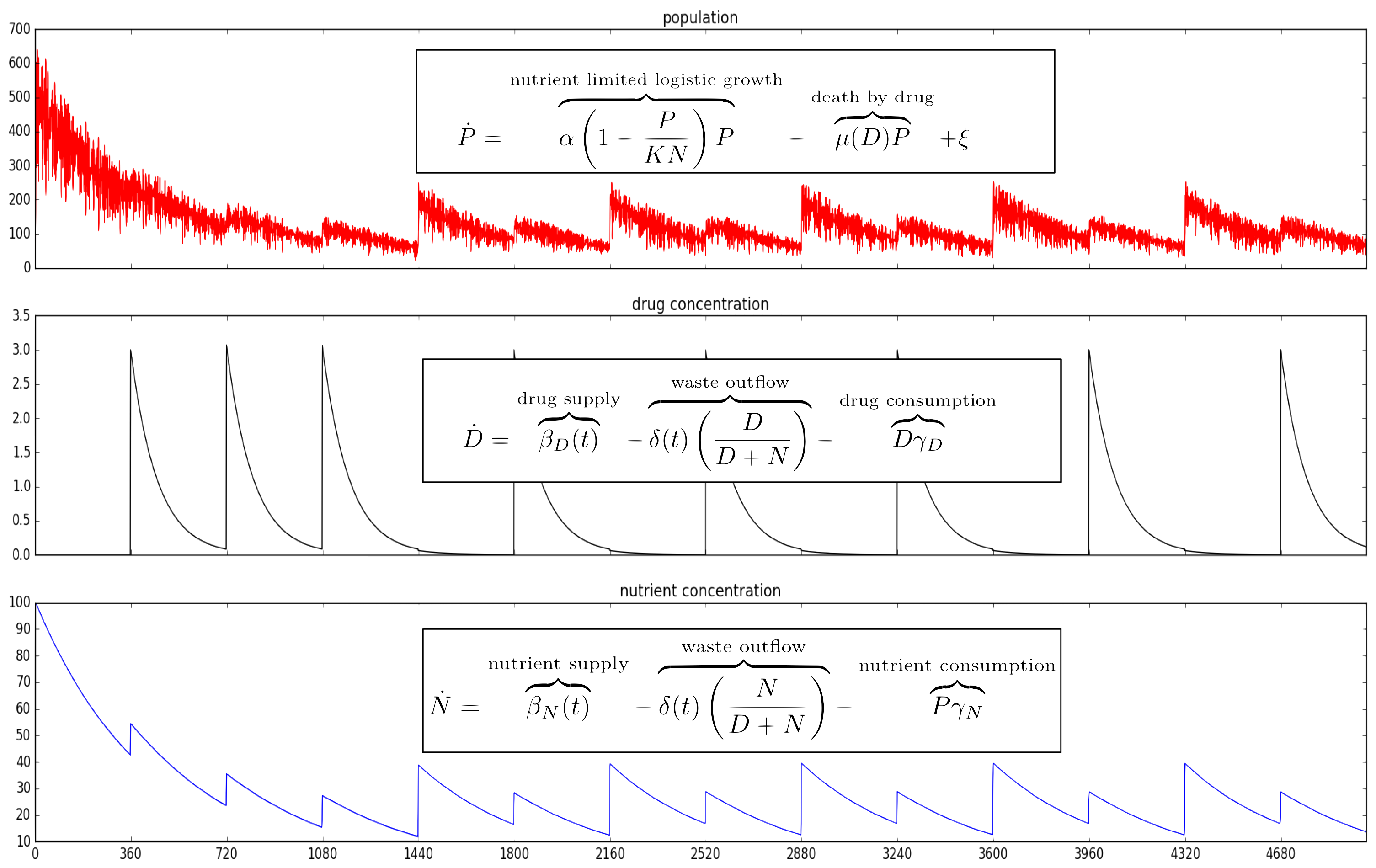
Mathematical model of proposed dynamics
Evolutionary Game Theory
We have an interest in the application of evolutionary game theory to the study of cancer, specifically to the evolution of resistance. Evolutionary game theory allows us to model and perturb the complex cooperative and competitive dynamics between sensitive tumor cells, resistant tumor cells, and the microenvironment. We use mathematical (systems of differential equations) and computational (numerical simulation) models (right), and an experimental game assay we have developed (co-culture and automated cell-counting) (left) to investigate these interactions.
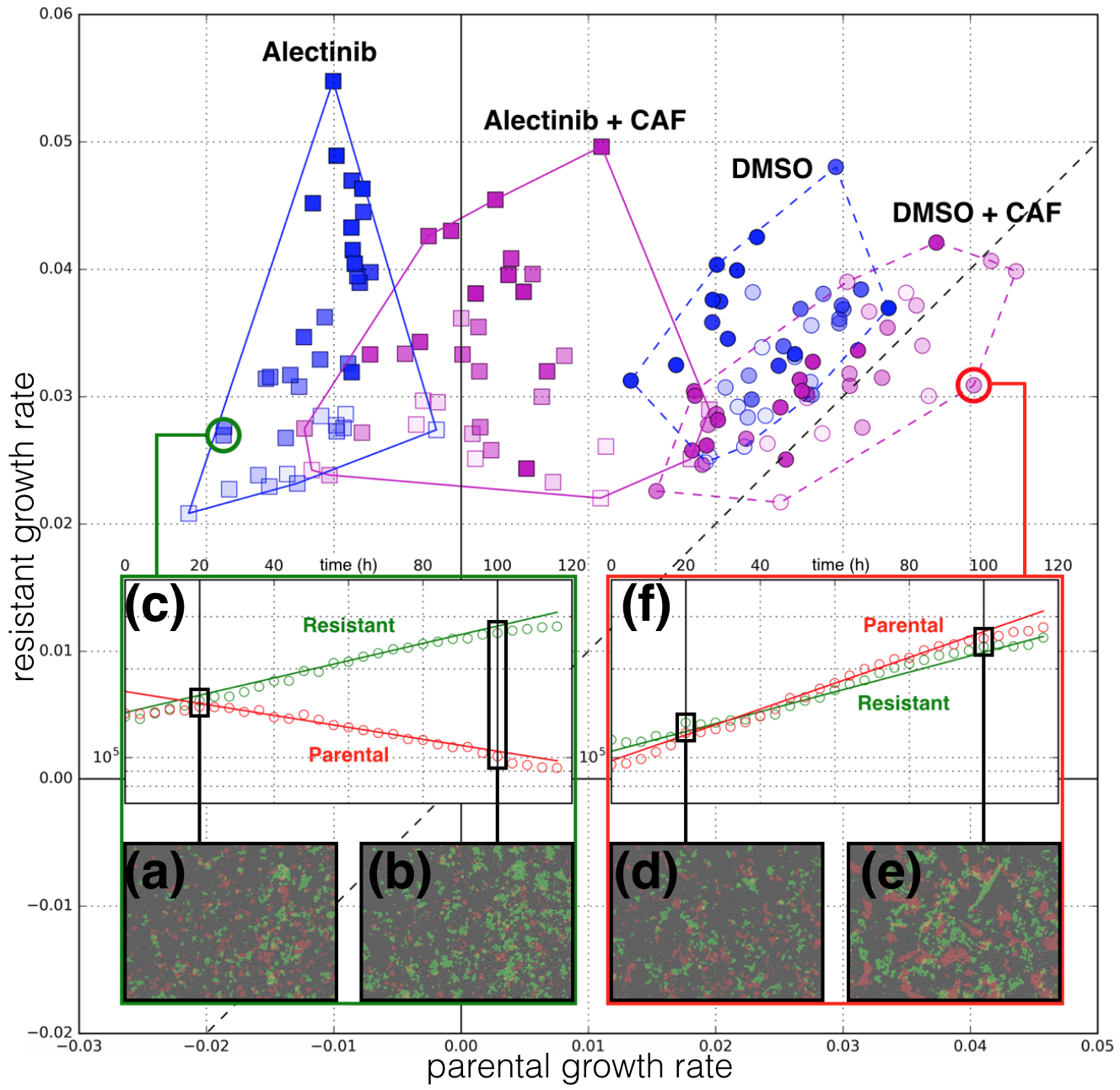
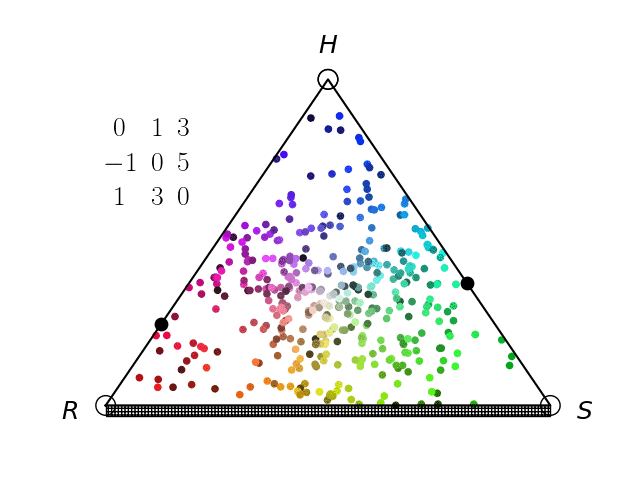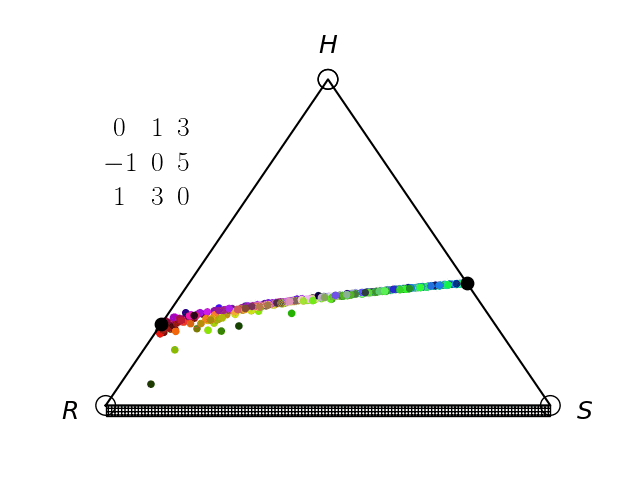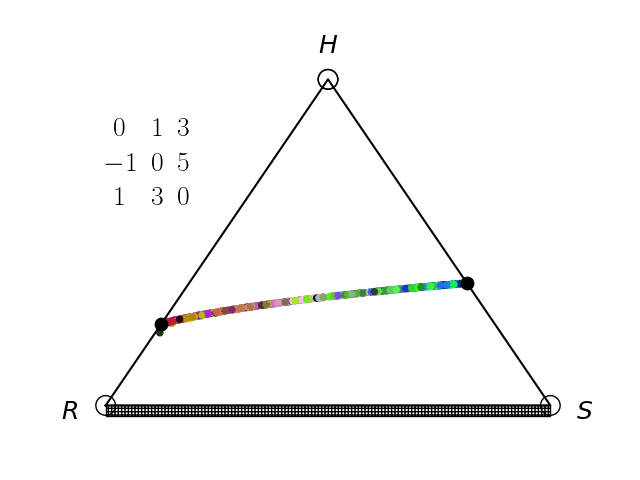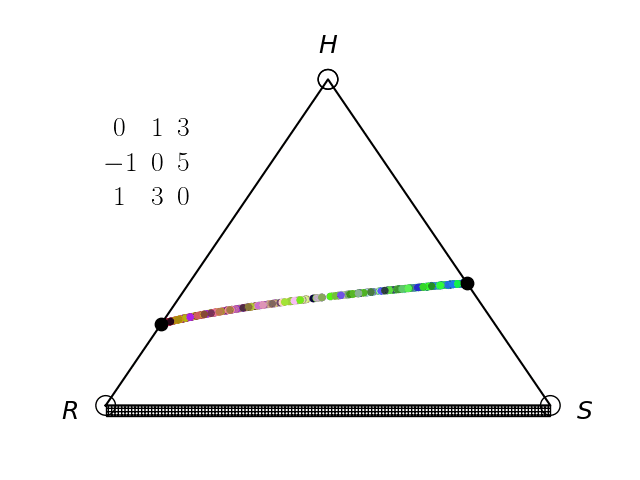
Data Science
With regards to data science, our interests span the breadth of clinical cancer research, ranging from developing novel biomarkers of disease, to selecting personalized therapies (including personalized radiation therapy dosing), and monitoring for resistance. Using datasets encompassing clinico-pathologic, biologic, and genomic data, we hope to design new tests, and lay the groundwork for novel anticancer therapies. To study large high dimensional datasets, we use a combination of high-performance computing and statistical approaches, on problems such as developing gene signatures for drug sensitivity and resistance in Ewings Sarcoma, understanding the functional role of newly characterized non-coding RNA, and determining methods of combining microRNAs into therapeutic cocktails.
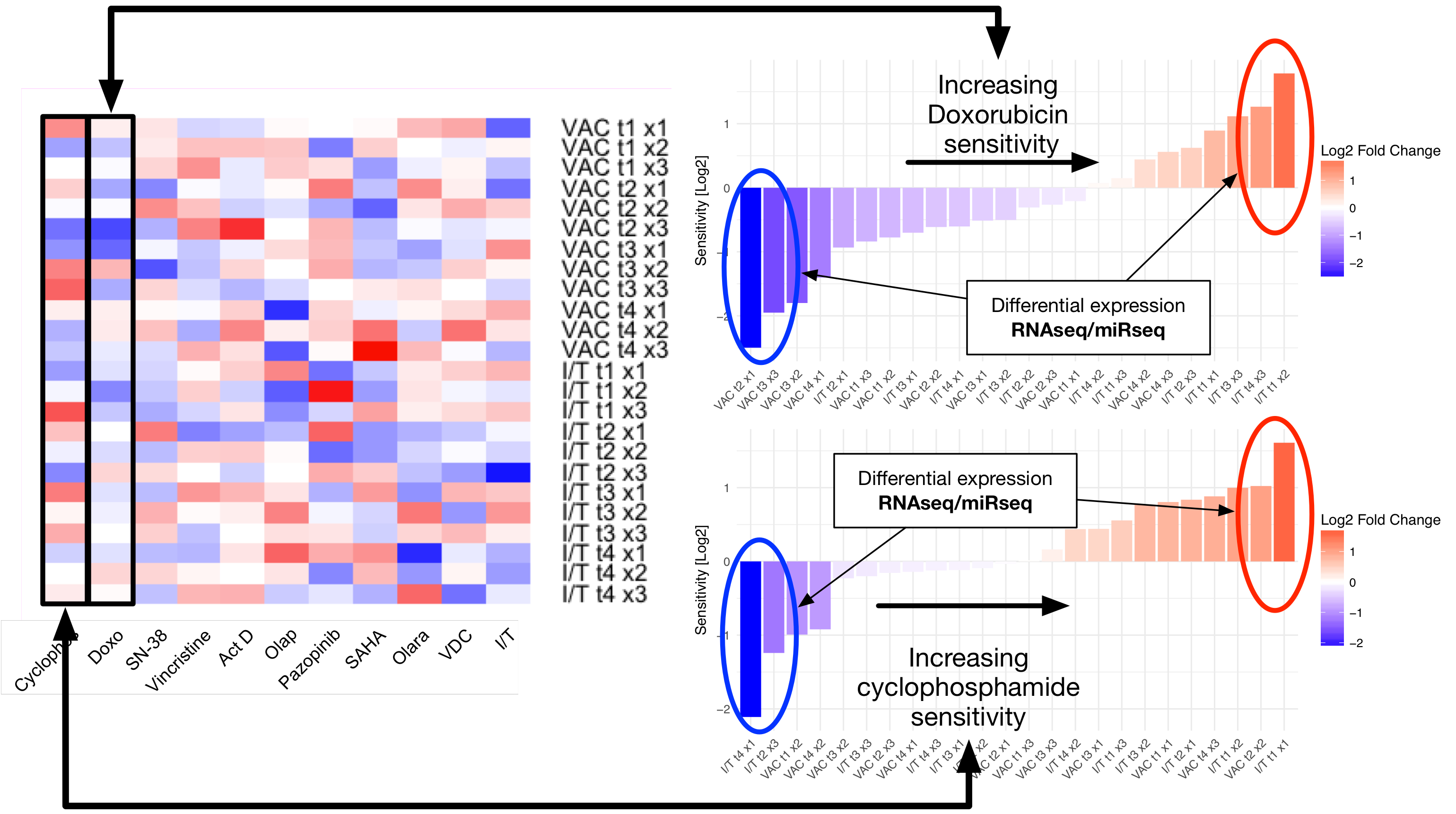
Our Team
Selected Publications
View publications for Jacob Scott, MD, DPhil
(Disclaimer: This search is powered by PubMed, a service of the U.S. National Library of Medicine. PubMed is a third-party website with no affiliation with Cleveland Clinic.)
1. Scott JG. Lee H, Barsoum WK, van den Bogert AJ. “The Effect of Tibiofemoral Loading on Proximal Tibiofibular Joint Motion.”Journal of Anatomy,November 2007;211(5):647-653 PMID: 17764523
2. Elias H , Scott JG, Metheny L, Quereshy F. “Multiple Myeloma Presenting as Mandibular Ill-defined Radiolucent Lesion with Numb Chin Syndrome: a case report.” Journal of Oral and Maxillofacial Surgery, September 2009;67(9):1991-1996 PMID: 19686938
3. Elias H , Scott JG, Metheny L, Quereshy F. “Multiple Myeloma Presenting as Mandibular Ill-defined Radiolucent Lesion with Numb Chin Syndrome: a case report.” Journal of Oral and Maxillofacial Surgery, September 2009;67(9):1991-1996 PMID: 19686938
4. OhS, Scott JG, Suh T, Kim S. “Measurement of Dose Discrepancies Due to Inhomogeneities and Radiographic Contrast in Balloon Catheter Brachytherapy.” Medical Physics, September 2009;36(9):3945-3954 PMID: 19810467
5. Packer CD, Carnell RC, Tomcho PM, Scott JG. “Development of a Four-Day Service-Learning Rotation for Third-Year Medical Students.” Teaching and Learning in Medicine, July 2010;22(3):224-228 PMID: 20563946
6. Scott JG, Tsai Y, Chinnaiyan P, Yu HM. “Effectiveness of Radiotherapy for Elderly Patients with Glioblastoma.” Int J Radiat Oncol Biol Phys. 2011 Sept 1;81(1):206-210 PMID: 20675068 *Highlighted as “Issue Highlight Article” September 2011
7. Basanta D✝, Scott JG✝, Rockne R, Swanson KR, Anderson ARA. “The role of IDH1 mutated tumour cells in secondary glioblastoma: an evolutionary game theoretical view.” Physical Biology. 2011 Feb; 8(1):015016. PMID: 21301070 *Top 3% of downloaded articles across IOP Journals for 2011
8. Scott JG, Suh JH, Elson P, Barnett GH, Vogelbaum MA, Peereboom DM, Stevens GH, Elinzano H, Chao ST. “Aggressive treatment is appropriate for glioblastoma multiforme patients 70 years or older: a retrospective review of 206 cases.” Neuro Oncology. 2011 Apr;13(4):428-3. PMID: 21363881 *Highlighted by Editorial - PMID: 21430110
9. Basanta D, Scott JG, Fishman MN, Ayala GE, Hayward SW, Anderson ARA. “Investigating prostate cancer tumour-stroma interactions - clinical and biological insights from an evolutionary game.” British Journal of Cancer, 3 Jan 2012; 106(1)1:174-181PMID: 22134510
10. Finkelstein SE, Rodriguez F, Dunn M, Scott JG, et al. “Knowing when to say when: prospective serial assessment of apoptosis, cell death, and lymphocyte infiltrates during the first clinical experience with intraprostatic autologous dendritic cell injection coordinated with radiation therapy.” Immunotherapy, 2012 Apr; 4(4):373-82 PMID: 22512631
11. Scott JG, Kuhn P, Anderson ARA. “Unifying Metastasis--Integrating intravasation, circulation and end organ colonization.” Nature Reviews Cancer 12, 445-446, Jul 2012 PMID: 22912952*Highlighted by Editorial - PMID: 23303865
12. Scott JG, Bauchet L, Fraum TJ, Nayak L, Cooper AR, Reiner AS, Chao ST, Suh JH, Vogelbaum MA, Peereboom DM, Zouaoui A, Mathieu-Daudé H, Fabbro-Peray P, Rigau V, Taillandier L, Abrey LE, DeAngelis LM, Shih JH, Iwamoto FM. “Recursive Partitioning Analysis Identifies Prognostic Groups for Glioblastoma Patients Aged 70 Years or Older.” Cancer, 15 Nov 2012;118(22): 5595-5600 PMID: 22517216
13. Scott JG, Basanta D, Anderson ARA, Gerlee P. “A mathematical model of tumor self-seeding reveals secondary metastatic deposits as drivers of primary tumor growth.” Journal of the Royal Society: Interface2013, Feb 20;10(82) DOI: 10.1098/rsif.2013.0011 PMID: 23427099
14. McTyre E✝, Scott JG✝, Chainnaiyan P. “Whole Brain Radiotherapy for Brain Metastases.” Surgical Neurology International. 2013 May 2;4(Supp 4):S236-44. DOI:10.4103/2152-7806.111301 http://goo.gl/4qugt PMID: 23717795
15. Belmonte-Beitia J, Woolley TE, Scott JG, Maini PK, Gaffney EA. “Modeling biological invasions: Individual to population scaling at interfaces.” Journal of Theoretical Biology 2013, Oct 7;334:1–12 DOI:10.1016/j.jtbi.2013.05.033 PMID: 23770401
16. Scott JG, Chinnaiyan P, Anderson ARA, Hjelmeland A, Basanta D. “Microenvironmental variables must influence intrinsic phenotypic parameters of cancer stem cells to affect tumourigenicity.” PLoS Computational Biology 2014, January 16 DOI: 10.1371/journal.pcbi.1003433 PMID: 24453958
17. Baldock AL†, Yagle K†, Born DE, Ahn S, Trister AD, Neal M, Johnston SK, Bridge C, Basanta D, Scott JG, Malone H, Sonabend AM, Canoll P, Mrugala MM, Rockhill JK, Rockne RC, and Swanson KR. “Invasion and proliferation kinetics in enhancing gliomas predict IDH1 mutation status.” Neuro Oncology 2014, Jun;16(6):779-786DOI: 10.1093/neuonc.nou027 *Highlighted by Editorial PMID: 24832620
18. Gallaher J, Cook LM, Gupta S, Araujo A, Dhillon J, Park JY, Scott JG, Pow-Sang J, Basanta D, Lynch CC. “Improving treatment strategies for patients with metastatic castrate resistant prostate cancer through personalized computational modeling.” Clinical and Experimental Metastasis. 2014 Dec;31(8):991-9. DOI: 10.1007/s10585-014-9674-1 PMID: 25173680
19. Scott JG, Fletcher AG, Maini PK, Anderson ARA, Gerlee P. “A filter-flow perspective of hematogenous metastasis offers a non-genetic paradigm for personalized cancer therapy” European Journal of Cancer. 2014 Nov;50(17):3068-75DOI:10.1016/j.ejca.2014.08.019 PMID: 25306188
20. Arora D, Frakes J, Scott JG, Opp D, Johnson C, Song J, Harris E. “Incidental radiation to uninvolved internal mammary lymph nodes in breast cancer.” Breast Cancer Research and Treatment. 2015 Jun;151(2):365-72 DOI:10.1007/s10549-015-3400-9 PMID: 25929764
21. Kaznatcheev A✝, Scott JG✝, Basanta D. “Edge effects in game theoretic dynamics of spatially structured tumours.” Journal of the Royal Society: Interface. 2015, 12:20150154. DOI:10.1098/rsif.2015.0154 PMID: 26040596
22. Nichol D, Jeavons P, Bonomo RA, Maini PK, Paul JL, Gatenby RA, Anderson ARA, Scott JG. “Steering Evolution with Sequential Therapy to Prevent the Emergence of Bacterial Antibiotic Resistance.” PLoS Computational Biology 2015 Sep 11: 11(9): e1004493. DOI:10.1371/journal.pcbi.1004493 PMID: 26360300
23. Scott JG, Fletcher AG, Anderson ARA, Maini PK. “Spatial metrics of tumour vascular organisation predict radiation efficacy in a computational model.” PLoS Computational Biology 2016, Jan 22: 12(1):e1004712 DOI: http://dx.doi.org/10.1101/029595 PMID: 26800503
24. Werner B, Scott JG, Sottoriva A, Anderson AR, Traulsen A, Altrock PM “The Cancer Stem Cell Fraction in Hierarchically Organized Tumors Can Be Estimated Using Mathematical Modeling and Patient-Specific Treatment Trajectories.” Cancer Research. 2016 Apr 1;76(7):1705-13 DOI: 10.1158/0008-5472.CAN-15-2069 PMID: 26833122
25. Naghavi AO, Gonzalez RJ, Scott JG, Mullinax JE, Abuodeh YA, Kim Y, Binitie O, Ahmed KA, Bui MM, Saini AS, Zager JS, Biagioli MC, Letson D, Harrison LB, Fernandez DC “Implications of staged reconstruction and adjuvant brachytherapy in the treatment of recurrent soft tissue sarcoma” Brachytherapy 2016 Jul: 15(4) 495-503 PMID: 27180128
26. Rietman EA, Scott JG, Tuszynski JA, Lakka-Klement G. “Personalized Anticancer Therapy Selection using Molecular Landscape Topology and Thermodynamics” Oncotarget 2016 Oct 26 DOI: http://dx.doi.org/10.18632/concotarget.12932 PMID: 27793055
27. Perni S, Mohamed AS, Scott JG, Enderling H, Garden AS, Gunn GB, Rosenthal DI, Fuller CD. CT-based volumetric tumor growth velocity: A novel imaging prognostic indicator in oropharyngeal cancer patients receiving radiotherapy. Oral Oncology 2016 Dec 31;63:16-22. DOI: http://dx.doi.org/10.1016/j.oraloncology.2016.10.022 PMID: 27938995
28. Grassberger C, Scott JG, and Paganetti H. "Biomathematical Optimization of Radiation Therapy in the Era of Targeted Agents." International Journal of Radiation Oncology Biology Physics 2017: 13-17. DOI: http://dx.doi.org/10.1016/j.ijrobp.2016.09.008PMID: 27979444
29. Scott JG, Berglund A, Schell M, Mihaylov I, Fulp WJ, Yue B, Welsh E, Caudell JJ, Ahmed K, Strom TS, Mellon E, Venkat P, Johnstone P, Foekens J, Lee J, Moros E, Dalton WS, Eschrich SA, McLeod H, Harrison LB, Torres-Roca JF. “A genomic framework for precision radiation therapy” Lancet Oncology 2017 Dec;18:2(202-211) DOI: http://dx.doi.org/10.1016/S1470-2045(16)30648-9 PMID: 27993569 *Highlighted by Editorial PMID: 27993570
30. Scott JG, Marusyk A. “Somatic clonal evolution: A selection-centric perspective” Biochimica Biophysica Acta - Reviews on Cancer- 2017 Apr 30;1867(2):139-50. PMID: 28161395
31. Kaznatcheev A, Vander Velde R, Scott JG, Basanta D. “Cancer treatment scheduling and dynamic heterogeneity in social dilemmas of tumour acidity and vasculature” British Journal of Cancer2017 March 14;116(6):785-792 doi: http://dx.doi.org/10.1101/067488 PMID: 28183139
32. Naghavi AO, Gonzalez RJ, Scott JG, Kim Y, Abuodeh YA, Strom TJ, Echevarria M, Mullinax JE, Ahmed KA, Harrison LB, Fernandez DC “Staged reconstruction brachytherapy has lower overall cost in recurrent soft-tissue sarcoma” Journal of Contemporary Brachytherapy 2017 Feb;9(1):20-29 doi: 10.5114/jcb.2017.65641
33. Naghavi AO, Fernandez DC, Mesko N, Juloori A, Martinez A, Scott JG, Shah C, Harrison LB. American Brachytherapy Society consensus statement for soft tissue sarcoma brachytherapy. Brachytherapy. 2017 Jun 30;16(3):466-89. doi: https://doi.org/10.1016/j.brachy.2017.02.004
34. Dhawan A, Nichol D, Kinose F, Abazeed M, Marusyk A, Haura E, Scott JG. “Collateral sensitivity networks reveal evolutionary instability and novel treatment strategies in ALK mutated non-small cell lung cancer” Scientific Reports2017 Apr 27;(1):1232 doi: http://dx.doi.org/10.1038/s41598-017-00791-8 PMID: 28450729
35. McFarland CD, Yaglom JA, Wojtkowiak JW, Scott JG, Morse DL, Sherman MY, Mirny LA. “Passenger DNA alterations reduce cancer fitness in cell culture and mouse models.” Cancer Research 2017 May 23.
36. Ottino-Loffler B, Scott JG, Strogatz SH. “Takeover times for a simple model of network infection” Physical Review E 2017 July 13;96(012313) doi: https://doi.org/10.1103/PhysRevE.96.012313
37. Zhou M, Scott JG, Chaudhury B, Hall L, Goldgof D, Yeom KW, Iv M, Ou Y, Kalpathy-Cramer J, Napel S, Gillies R. Radiomics in Brain Tumor: Image Assessment, Quantitative Feature Descriptors, and Machine-Learning Approaches. American Journal of Neuroradiology. 2017 Oct 5.
38. Karthik N, Juloori A, Mesko N, Scott JG, Shah CS. “Factors Associated with Acute and Chronic Wound Complications in Patients with Soft-Tissue Sarcoma with Long-Term Follow-up” - in press, American Journal of Clinical Oncology
39. Ottino-Loffler B, Scott JG, Strogatz SH. “Evolutionary dynamics of incubation periods” eLife 2017;6:e30212 doi: 10.7554/eLife.30212
40. Yoon N, Vander Velde R, Marusyk A, Scott JG. “Optimal therapy scheduling based on a pair of collaterally sensitive drugs.” -in press, Bulletin of Mathematical Biology preprint: https://www.biorxiv.org/content/early/2017/10/02/196824
41. Kaznatcheev A, Peacock J, Basanta D, Marusyk A, Scott JG. "Cancer associated fibroblasts and alectinib switch the evolutionary games that non-small cell lung cancer plays” - in revisions, Nature Ecology and Evolution pre-print: http://www.biorxiv.org/content/early/2017/08/21/179259
42. Alfonso JCL, Parsai S, Joshi N, Godley A, Koyfman S, Caudell JJ, Fuller CD, Enderling H, Scott JG. “Temporally-Feathered Intensity Modulated Radiation Therapy: A technique to reduce normal tissue toxicity” - in revisions, Medical Physics pre-print: http://www.biorxiv.org/content/early/2017/09/03/184044
43. Nichol D, Rutter J, Bryant C, Jeavons P, Anderson ARA, Bonomo RA, Scott JG. “Collateral sensitivity is contingent on the repeatability of evolution” - in revisions, Nature Communications pre-print: http://www.biorxiv.org/content/early/2017/09/07/185892
44. Dhawan A, Barberis A, Cheng W, Domingo E, West C, Maughan T, Scott JG, Harris AL, Buffa FM. “sigQC: A procedural approach for standardising the evolution of gene signatures.” - in revisions, Nature Protocols preprint: https://www.biorxiv.org/content/early/2017/10/16/203729
45. Ahmed KA, Scott JG, Arrington JA, Naghavi AO, Grass GD, Perez BA, Caudell JJ, Berglund AE, Welsh EA, Eschrich SA, Dilling TJ, Torres-Roca JF. “Radiosensitivity of Lung Metastases by Primary Histology and Implications for Stereotactic Body Radiotherapy Using the Genomically Adjusted Radiation Dose”. - in press, Journal of Thoracic Oncology.
46. Scott JG, Dhawan A, Hjelmeland A, Lathia J, Chumakova A, Hitomi M, Fletcher AG, Maini PK, Anderson ARA. “Recasting the cancer stem cell hypothesis: unification using a continuum model of microenvironmental forces.” preprint: https://www.biorxiv.org/content/early/2017/08/14/169615
47. Dhawan A, Harris A, Buffa FM, Scott JG. “Endogenous miRNA sponges mediate the generation of oscillatory dynamics for a non-coding RNA network.” preprint: https://www.biorxiv.org/content/early/2018/03/31/292029
Careers
We are always looking for great folks to work with!
As the primary goal of our lab is to come up with innovative solutions to hard problems, we are interested in scientists with diverse backgrounds and training. We currently have systems biologists, mathematicians, bioinformaticians, physicists, computer scientists, physicians and a cell biologist.
Graduate Students
I am a certified trainer at Case Western Research University in the Systems Biology and Medical Scientist Training Programs, but accept students from other disciplines as well (math/physics/biology). Interested PhD students should contact Jake with specific questions.
Postdocs
We are always looking for passionate and creative postdocs. We have a slight preference for computationally trained folks, but wouldn't rule out somoene with strong evolutionary or cell biological training who was interested in learning some computation or engineering. If you are interested in working with us, please email Jacob with your CV, a brief description of your background, and your goals in pursuing a postdoc.
Internships, Undergraduates & Visitors
We are always interested in hosting visitors, but time and space are limited, so must choose carefully based on current human capital. Please reach out with questions.
Lab Values & Expectations
Work hard, take care of yourself and others, and be nice.
Diversity & Inclusion Statement
Our lab is committed to creating and maintaining an environment in which all are welcome and respected - one that is inclusive of race, gender, faith, sexual orientation, ability, and socioeconomic status. We acknowledge that many identities are underrepresented in STEM and in academia, and that systemic racism and other discriminations create obstacles for students and postdocs. We strongly condemn behaviors and institutions that perpetuate these inequities, and we as a lab must act to dismantle the structural and systemic barriers to higher education.
Mission Statement
The mission of our lab is to create a space for students and postdocs to develop the skills necessary to be able to effectively question, research, and communicate scientific topics. We understand that differences will exist in the level of training, knowledge, personal circumstances, and life and career goals of lab members. As such, our goal is to provide an equitable academic opportunity while taking these conditions into account.
Training at Lerner Research Institute
Our education and training programs offer hands-on experience at one of the nationʼs top hospitals. Travel, publish in high impact journals and collaborate with investigators to solve real-world biomedical research questions.
Learn MoreResearch News

Understanding how evolution drives resistance to endometrial cancer chemotherapy can help researchers force tumors to evolve vulnerabilities to new treatments.

A new study identifies evolutionary dynamics that support pre-existing treatment resistance mutations in lung cancer and beyond.

Researchers used reinforcement learning to design antibiotic regimens to prevent treatment resistance.

The new Cleveland Clinic-developed model incorporates how evolution contributes to drug effectiveness and dosage.

Work with computer and preclinical models aims to bridge the gap between studying evolutionary biology in the lab and delivering to patient care.

Understanding cell growth dynamics may be the key to controlling therapeutic resistance.

Dr. Scott invented a new radiation planning technique called Temporally Feathered Radiation Therapy to reduce toxicity, which has now been shown to be safe and feasible in a small clinical study.

Dr. Scott and colleagues validated the genomic-adjusted radiation dose to be beneficial as a pan-cancer predictor of radiation therapy efficacy in new clinical study.

Dr. Card, a postdoctoral fellow in the Scott lab, has been named a 2020 Hanna Gray Fellow, a fellowship that helps provide support for underrepresented and early-career biomedical researchers.

Rather than a one-size-fits-all approach, new technology from Dr. Scott provide opportunity to choose personalized radiation dose to improve outcomes and reduce toxicity.