Emily Zabor Research Program
-
Emily Zabor Research Program
- Principal Investigator
- Research
- Our Team
- Publications
- Careers
- Research News

Emily Zabor, DrPH
Associate Staff
Joint Appointment in Taussig Cancer Institute
Email: [email protected]
Location: Cleveland Clinic Main Campus
Research
The Emily Zabor research program studies statistical methods for oncology studies, including clinical trial design, retrospective data analyses, and survival analyses. In addition to methodologic development, statistical applications are conducted across the range of oncology, including both solid tumors and hematologic malignancies. The primary area of statistical methods research is in early phase oncology clinical trial design. As the treatment landscape in oncology shifts away from traditional cytotoxic therapies and towards a variety of types of biomarker-targeted therapies, there is a need for innovation in statistical methods that are tailored to the context of these novel therapeutics. Both Bayesian and frequentist statistical paradigms are employed. The secondary area of statistical methods research is in retrospective data analysis methods, with a focus on survival analysis. As data from electronic health records becomes more extensive and accessible, statistical methods that account for the implicit biases in analyses based on such data are needed.
Biography
Emily Zabor, DrPH is an Associate Staff Biostatistician at the Cleveland Clinic in the Department of Quantitative Health Sciences, with a joint appointment in the Taussig Cancer Institute. Emily also holds an academic appointment as Assistant Professor of Medicine at the Cleveland Clinic Lerner College of Medicine of Case Western Reserve University. Prior to joining the faculty at Cleveland Clinic, Emily worked for nine years as a research biostatistician at Memorial Sloan Kettering Cancer Center in New York City. Emily holds an MS degree in biostatistics from The University of Minnesota and a DrPH degree in biostatistics from Columbia University.
Education & Professional Highlights
DrPH, Biostatistics - Columbia University, New York, NY
MS, Biostatistics - University of Minnesota, Minneapolis, MN
BA, Anthropology - Grinnell College, Grinnell, IA
Research
Early Phase Oncology Clinical Trial Design
Phase I oncology clinical trials are increasingly adding dose expansion cohorts following the initial dose escalation phase. But the dose expansion cohorts are not always planned in advance, so can be subject to poor statistical properties and at times much too large sample size. I have proposed two criteria for selecting a Bayesian predictive probability design for futility monitoring in order to improve efficiency in dose expansion cohort design. The proposed method evaluates a grid of posterior and predictive thresholds, and evaluates them for the frequentist operating characteristics of type I error and power. The optimal accuracy or optimal efficiency designs can be used to select from among the various design options, as depicted in the below figure. Both designs tend to result in a trial design with improved power and lower average sample size under both the null and alternative as compared to a traditional single interim look design.
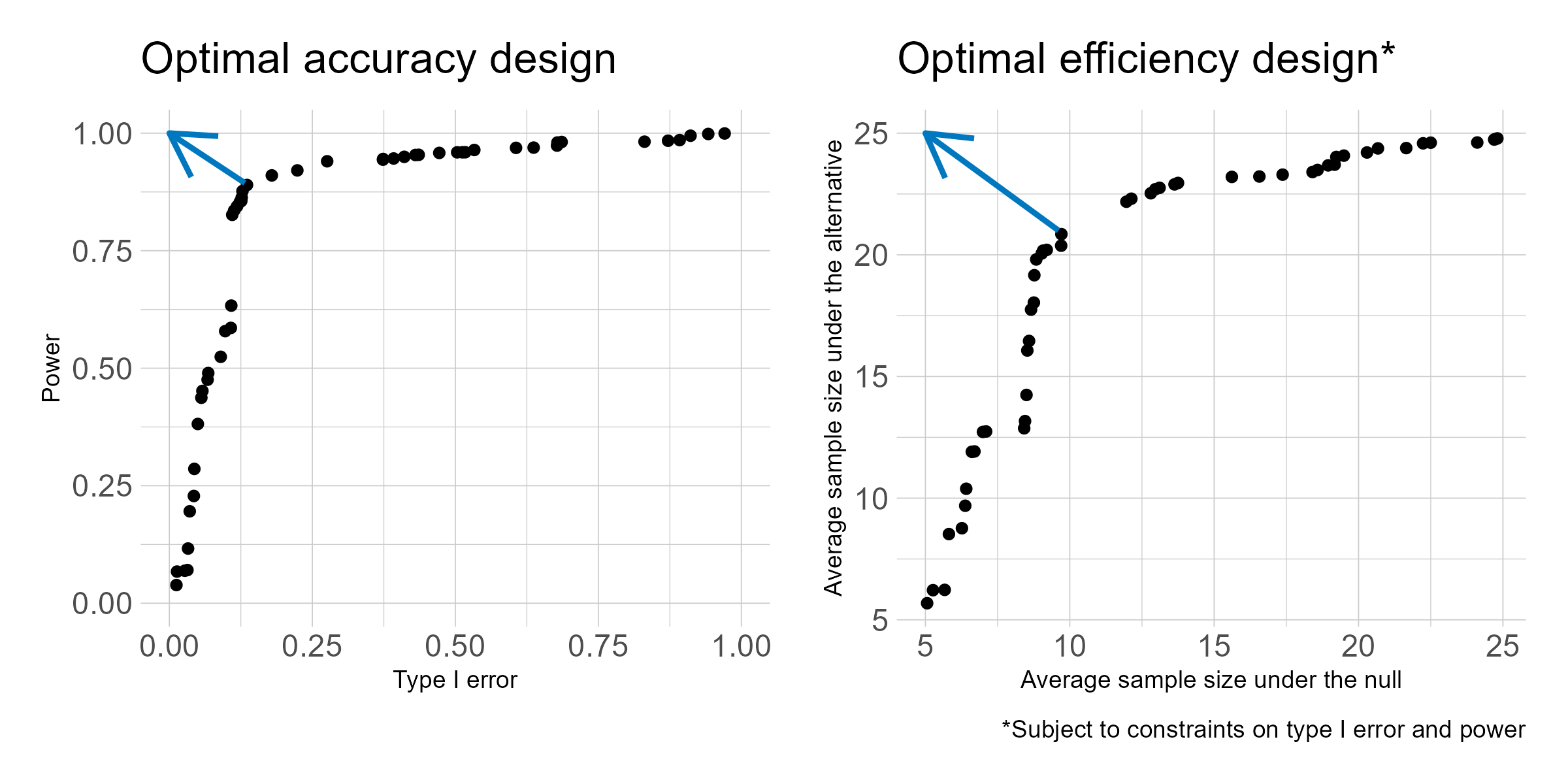
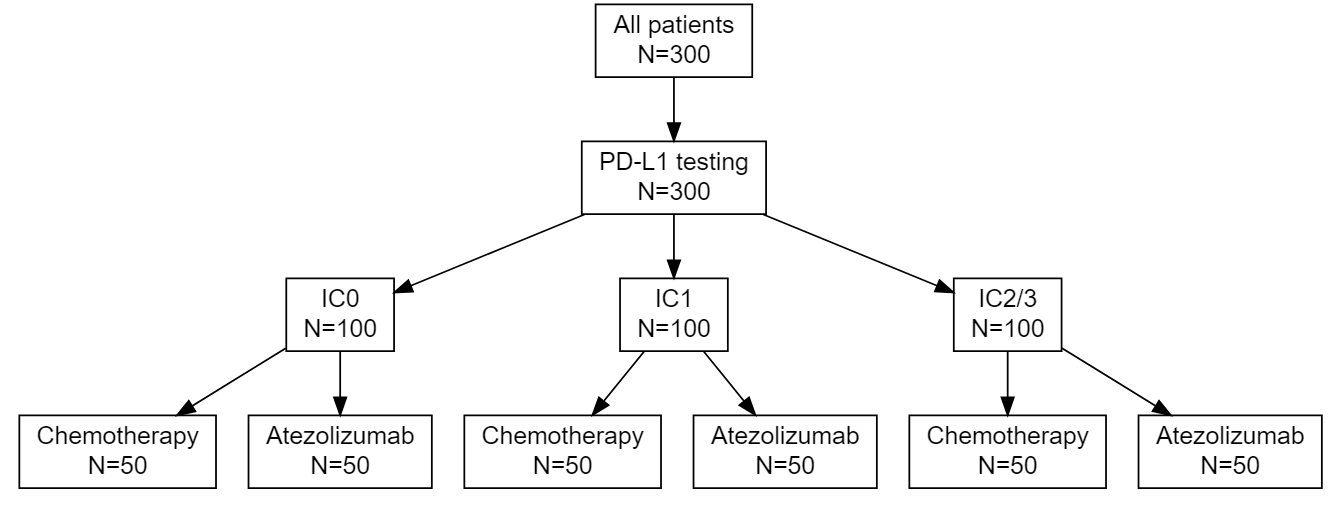
As the treatment landscape in oncology shifts away from the traditional cytotoxic treatments and toward a variety of biomarker-targeted therapies, the historical control rates typically used in single arm phase II trials, which arise as population averages, are not always appropriate. I proposed three designs for randomized phase II trials using Bayesian predictive probability for futility monitoring, based on the optimal accuracy and optimal efficiency designs described above. An example of the stratified randomization design schema is shown below. This design would be well suited to the context of a biomarker that is hypothesized to be prognostic, as it provides information on the control response rate within each biomarker-stratified subgroup. Alternative designs include a pooled control arm design and an enrichment design. A design can be selected based on the specific clinical and therapeutic context.
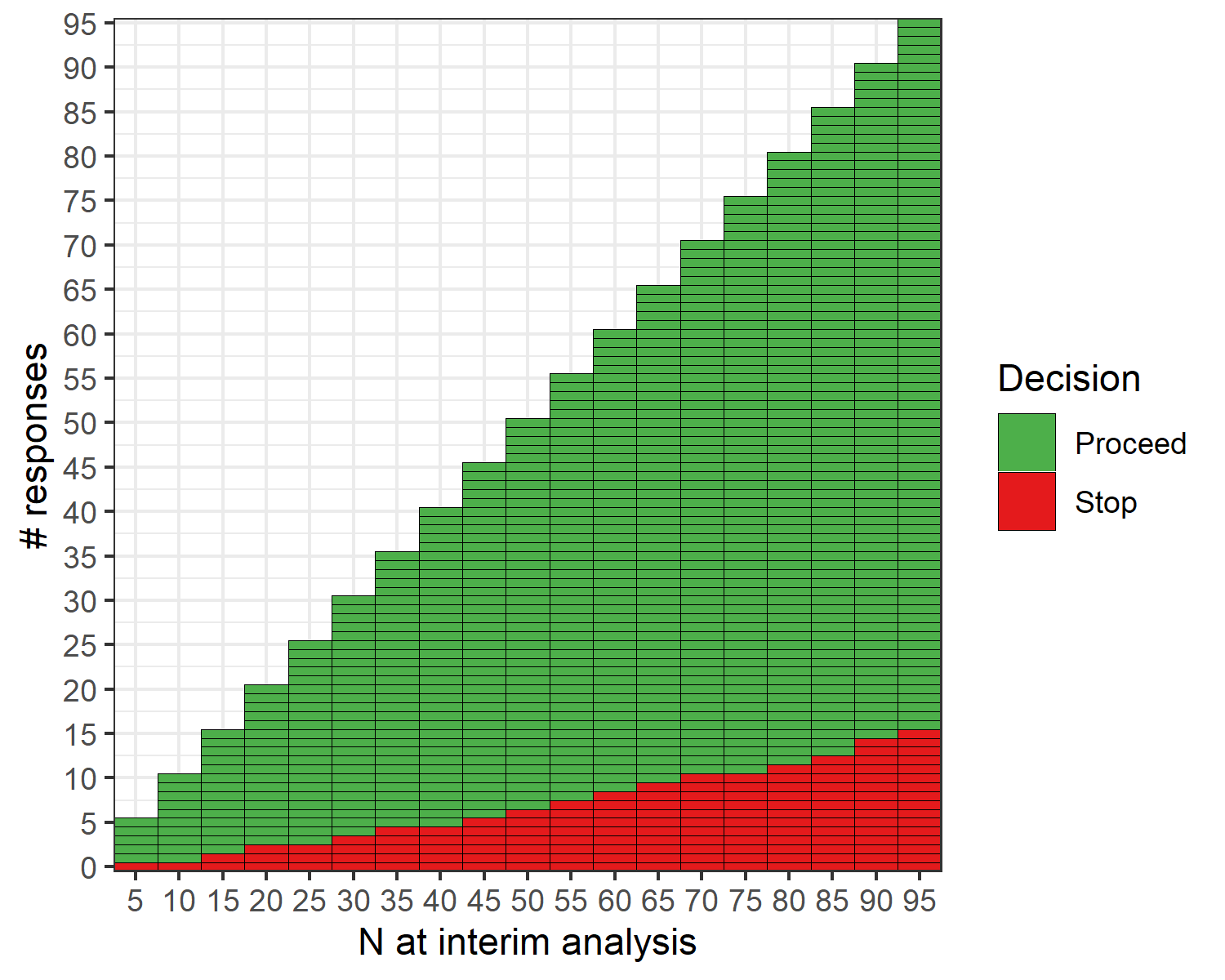
The open-source R software package {ppseq} was developed to implement the described methods, and details can be found at https://www.emilyzabor.net/ppseq/. The proposed methodology is implicitly computationally intensive due to the need for simulation-based methods to evaluate designs across a grid of posterior and predictive thresholds. The {ppseq} package can produce both a decision table and a decision plot, as illustrated in the below example, that can be obtained at the trial design stage to allow for seamless trial conduct without the need for any interim computations. At each interim monitoring point, depicted on the x-axis, you can see the number of responses required to continue the trial and the number of responses that would stop the trial early on the y-axis.
Retrospective Data Analysis Methods
Electronic health records data are becoming more extensive and also more easily accessible. As a result, retrospective data analyses to answer important clinical questions can be conducted with relative ease. In oncology, survival endpoints such as progression-free survival and recurrence-free survival are common. But these endpoints are subject to particular sources of bias and mis-specification, which compound in the context of data from retrospective sources. This area of statistical methods research is driven by problems that arise in collaborative projects analyzing retrospective survival endpoints. One example is in endpoints that are subject to interval censoring. When patients come in for scheduled visits to follow-up on their cancer treatment, a recurrence could be detected. But the true recurrence date will always be unknown, it will only be known that the patient had a recurrence between the date of their last clear scan, and the date on which the recurrence was detected.
 Specialized statistical methods are required to analyze this type of data, as the common approach of using the date on which the recurrence was detected as the true recurrence date can introduce bias under certain conditions. In recent research we found that when there are differential visit rates between two comparison groups of interest, there is increasing bias in the standard analysis approach as the difference in visit frequency increases. Using the Cox regression model for interval censored data with censoring on the date a patient was last known to be disease-free can return unbiased estimates.
Specialized statistical methods are required to analyze this type of data, as the common approach of using the date on which the recurrence was detected as the true recurrence date can introduce bias under certain conditions. In recent research we found that when there are differential visit rates between two comparison groups of interest, there is increasing bias in the standard analysis approach as the difference in visit frequency increases. Using the Cox regression model for interval censored data with censoring on the date a patient was last known to be disease-free can return unbiased estimates.
Our Team
Selected Publications
A comprehensive list of publications can be found on Google Scholar or via a PubMed search. A selection of recent publications is also included below.
Selected recent publications:
- Eaton, A. A.*, & Zabor, E. C.* (2022). Analysis of composite endpoints with component-wise censoring in the presence of differential visit schedules. Stat Med. doi:10.1002/sim.9312 *equally contributing authors
- Hobbs, B. P., Pestana, R. C., Zabor, E. C., Kaizer, A. M., & Hong, D. S. (2022). Basket Trials: Review of Current Practice and Innovations for Future Trials. J Clin Oncol, Jco2102285. doi:10.1200/jco.21.02285
- Lemmon, C. A., Zabor, E. C., & Pennell, N. A. (2022). Modeling the Cost-Effectiveness of Adjuvant Osimertinib for Patients with Resected EGFR-mutant Non-Small Cell Lung Cancer. Oncologist, 27(5), 407-413. doi:10.1093/oncolo/oyac021
- Sittenfeld, S., Zabor, E. C., Hamilton, S. N., Kuerer, H. M., El-Tamer, M., Naoum, G. E., Truong, P. T., Nichol, A., Smith, B. D., Woodward, W. A., Moo, T. A., Powell, S. N., Shah, C. S., Taghian, A. G., Abu-Gheida, I., & Tendulkar, R. D. (2022). A multi-institutional prediction model to estimate the risk of recurrence and mortality after mastectomy for T1-2N1 breast cancer. Cancer, 10.1002/cncr.34352. Advance online publication. https://doi.org/10.1002/cncr.34352
- Zabor, E. C., Kane, M. J., Roychoudhury, S., Nie, L., & Hobbs, B. P. (2022). Bayesian basket trial design with false-discovery rate control. Clin Trials, 19(3), 297-306. doi:10.1177/17407745211073624
- Zabor, E. C., Kaizer, A. M., Garrett-Mayer, E., & Hobbs, B. P. (2022). Optimal Sequential Predictive Probability Designs for Early-Phase Oncology Expansion Cohorts. JCO Precis Oncol, 6, e2100390. doi:10.1200/po.21.00390
- Zabor, E. C., Raval, V., Luo, S., Pelayes, D. E., & Singh, A. D. (2022). A Prediction Model to Discriminate Small Choroidal Melanoma from Choroidal Nevus. Ocul Oncol Pathol, 8(1), 71-78. doi:10.1159/000521541
- Zabor, E. C., Reddy, C. A., Tendulkar, R. D., & Patil, S. (2022). Logistic Regression in Clinical Studies. Int J Radiat Oncol Biol Phys, 112(2), 271-277. doi:10.1016/j.ijrobp.2021.08.007
- Zabor, E. C., Kaizer, A. M., & Hobbs, B. P. (2020). Randomized Controlled Trials. Chest, 158(1s), S79-s87. https://doi:10.1016/j.chest.2020.03.013
Careers
Postdoctoral researcher in cancer biostatistics
The Emily Zabor lab is seeking a postdoctoral researcher in cancer biostatistics to join the Department of Quantitative Health Sciences at Cleveland Clinic.
About the position
The main focus of the postdoctoral researcher will be the development of statistical methods for early phase oncology clinical trials. The postdoctoral researcher will be mentored in a variety of professional and analytic skills that will serve them in their early career as a biostatistician including independent statistical methods research, simulation studies, the oral and written communication of scientific results, and conducting effective scientific collaborations. In addition to publishing manuscripts, the postdoctoral researcher will have the opportunity to contribute to R package development. The postdoctoral researcher will also have the opportunity to work on collaborative projects in a variety of oncology applications, such as development of prediction models, cost-effectiveness analyses, survival modeling, and analysis of electronic health records data. Experience or interest in Bayesian statistics and R programming would be useful for this position.
Details
Cleveland Clinic provides a comprehensive benefits package that includes paid time off, parental leave, annual travel stipend, and health care benefits. Additional details of the included benefits are available on request. The salary range for this position is in line with current NIH postdoctoral pay. The position is for up to 2 years, depending on satisfactory performance in the first year. Start date is flexible and the position will remain open until filled.
To apply
Interested applicants should send 1) a letter of interest describing their research interests, relevant skills and experience, and career goals, 2) their most up-to-date CV, and 3) the names and contact information for 2-3 references via e-mail to Emily Zabor at [email protected]. All applications will be considered thoroughly and equally.
Training at Lerner Research Institute
Our education and training programs offer hands-on experience at one of the nationʼs top hospitals. Travel, publish in high impact journals and collaborate with investigators to solve real-world biomedical research questions.
Learn MoreResearch News

The model uses a patient’s records to estimate their gastric cancer risk and flag individuals who need preventative stomach cancer screening.